MolGPT#
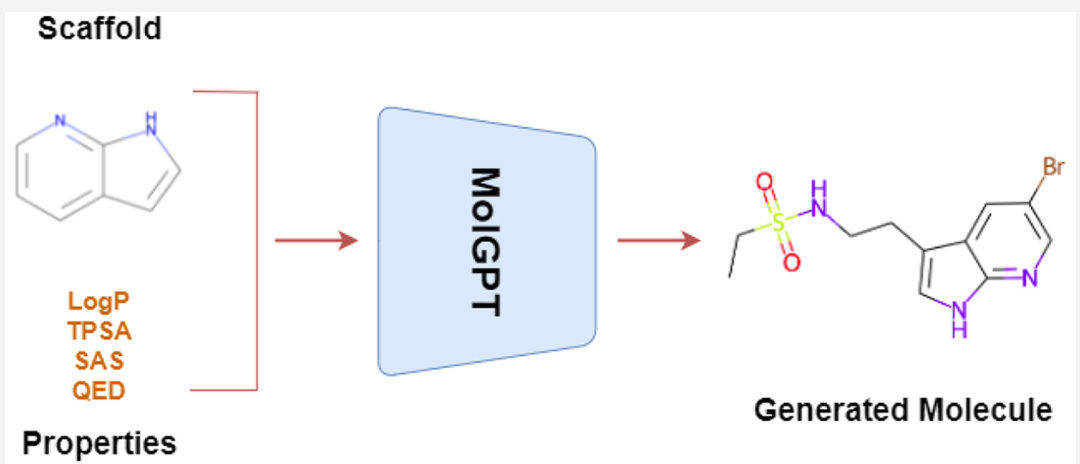
1. Methods#
The input of the model
Molecular SMILES
Scaffold
Properties:\(\left\{\begin{aligned}&\log{P}\quad \text{ ( The logarithm of the partition coefficient.)}\\ &SAS \quad \text{ (Synthetic Accessibility score) }\\&TPSA \quad \text{ (Topological Polar Surface Area)}\\&QED \quad \text{ (Quantitative Estimate of Drug-likeness )}\end{aligned}\right.\)
Dataset used
MOSES or the GuacaMol
**MOSES: **A data set composed of 1.9 million clean lead-like molecules from the Zinc data set46 with molecular weight ranging from 250 to 350 Da, number of rotatable bonds lower than 7, and XlogP below 3.5.
**GuacaMol: **A subset of the ChEMBL 24 database that contains 1.6 million molecules.
1.1. The architecture of Transformer#
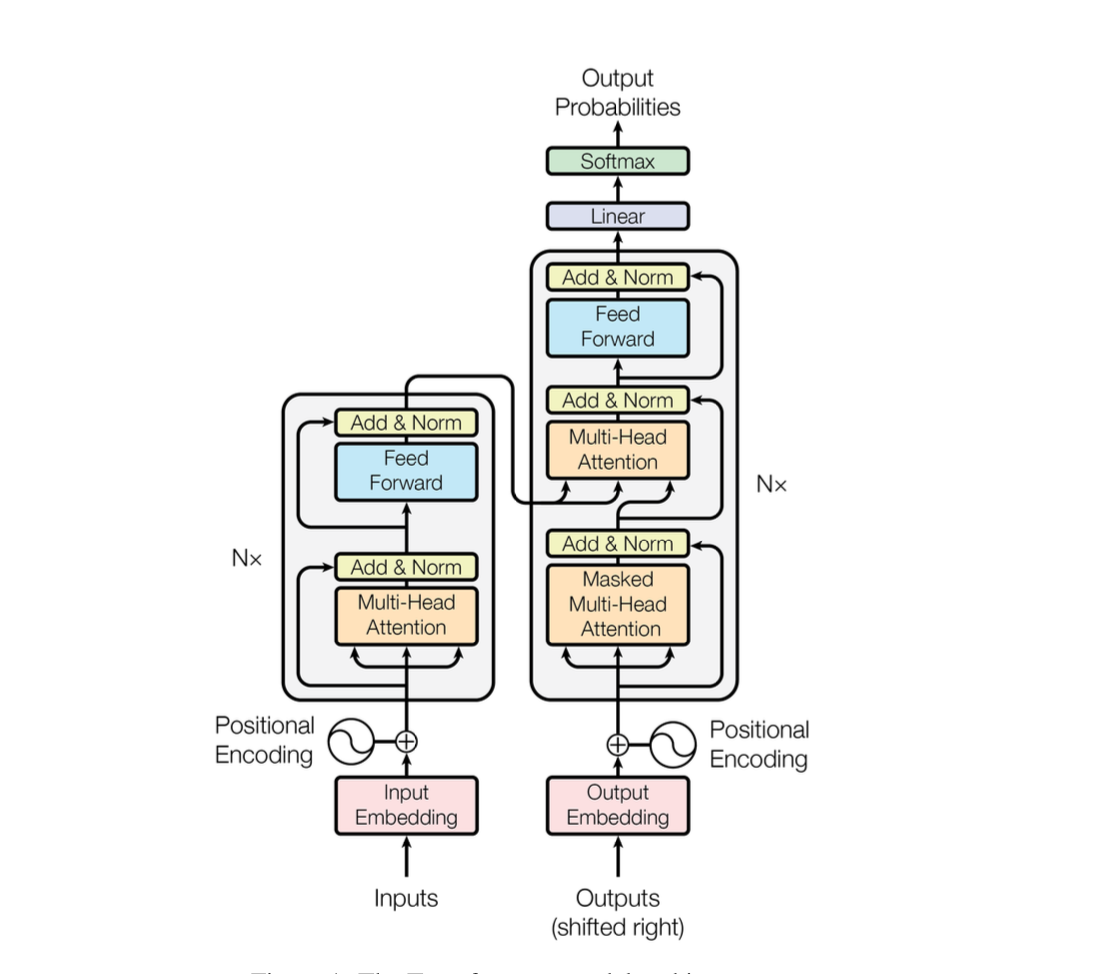
Detailed tutorial can be seen in: Transformer.md
2. Training Procedure and Evaluation Metrics#
Model is trained for 10 epochs using the Adam optimizer with a learning rate of \(6 \times 10^{-4}\).
Evaluation metrics :
Validity: The fraction of a generated molecules that are valid.
Uniqueness: The fraction of valid generated molecules that are unique.
**Novelty: **The fraction of valid unique generated molecules that are not in the training set.
Internal Diversity (\(\mathbf{IntDiV_p}\) ): Measures the diversity of the generated molecules.
**Freshet ChemNet Distance(FCD): **Calculated using the features of the generated molecules and the features of molecules in the data set.
KL Divergence: KL divergence between two distributions P and Q for any given property is a measure of how well Q approximates P
3. RESULTS AND DISCUSSION#
Nonconditioned Molecular Generation.
Generation-based on Single and Multiple Properties.
Generation Based on Scaffold.
Generation Based on Scaffold and Property.